Tracking centroids of segmented cells
[ ]:
%pip install -q --upgrade -r requirements.txt
Importing packages
laptrack.LapTrack is the core object for tracking.
We also import regionprops_table from skimage to calculate the centroids of the segmentation masks.
[1]:
import napari
import numpy as np
import pandas as pd
from matplotlib import pyplot as plt
from skimage.measure import regionprops_table
from laptrack import LapTrack
from laptrack import datasets
from laptrack.data_conversion import convert_split_merge_df_to_napari_graph
Load images and showing in napari
First, we load images to the napari viewer.
[2]:
viewer = napari.Viewer()
images, labels = datasets.cell_segmentation()
viewer.add_image(images)
labels_layer = viewer.add_labels(labels)
Calculating label centroids by regionprops_table
We then calculate the centroids for each labeled region in each frame.
[3]:
regionprops = []
for frame, label in enumerate(labels):
df = pd.DataFrame(regionprops_table(label, properties=["label", "centroid"]))
df["frame"] = frame
regionprops.append(df)
regionprops_df = pd.concat(regionprops)
Tracking centroids by LapTrack
This is where we use LapTrack. We track the centroids of the segmented regions.
[4]:
lt = LapTrack(track_cost_cutoff=15**2, splitting_cost_cutoff=30**2)
track_df, split_df, merge_df = lt.predict_dataframe(
regionprops_df.copy(),
coordinate_cols=["centroid-0", "centroid-1"],
only_coordinate_cols=False,
)
[5]:
track_df = track_df.reset_index()
graph = convert_split_merge_df_to_napari_graph(split_df, merge_df)
viewer.add_tracks(
track_df[["track_id", "frame", "centroid-0", "centroid-1"]],
graph=graph,
)
[5]:
<Tracks layer 'Tracks' at 0x7f8ec8bcbc10>
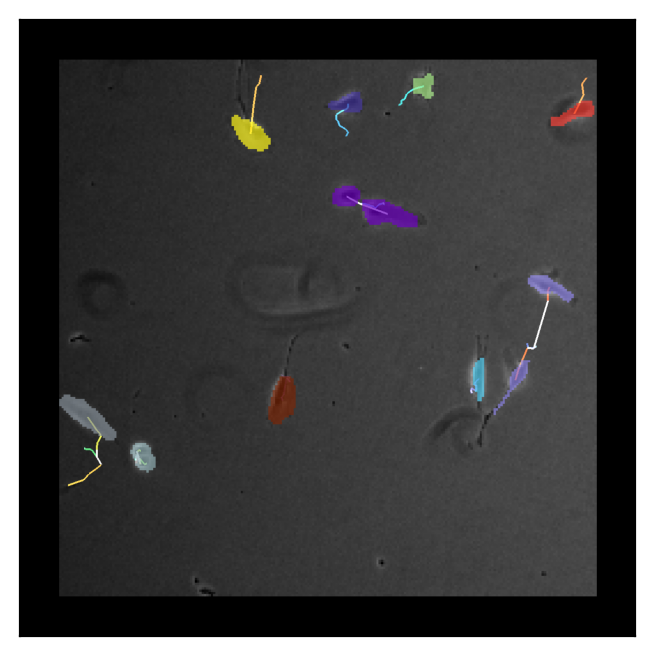
Showing clonal cells by the same colors
To check the result, we show the clonal cell regions by the same colors.
[6]:
new_labels = np.zeros_like(labels)
for i, row in track_df.iterrows():
frame = int(row["frame"])
inds = labels[frame] == row["label"]
new_labels[frame][inds] = int(row["tree_id"]) + 1
viewer.add_labels(new_labels)
labels_layer.visible = False
[7]:
viewer.dims.current_step = (9, 0, 0)
plt.imshow(viewer.screenshot())
plt.xticks([])
plt.yticks([])
[7]:
([], [])
[ ]: