Core tracking API by examples
This notebook illustrates the use of core APIs with a simple example.
[1]:
try:
import google.colab
%pip install -q --upgrade laptrack matplotlib spacy flask pandas
# upgrade packages to avoid pip warnings if the notebook is run in colab
except:
%pip install -q --upgrade laptrack matplotlib pandas
Note: you may need to restart the kernel to use updated packages.
Note: Restart the runtime when you are on Google Colab and you see an error in matplotlib.
Importing packages
laptrack.LapTrack is the core object for tracking.
[2]:
from os import path
import pandas as pd
from IPython.display import display
from matplotlib import pyplot as plt
from laptrack import LapTrack
from laptrack import datasets
plt.rcParams["font.family"] = ""
Matplotlib is building the font cache; this may take a moment.
Loading data
Load the example point coordinates from a CSV file.
spots_df has columns ["frame", "position_x", "position_y"] (can be arbitrary, and the column names for tracking will be specified later).
[3]:
spots_df = datasets.simple_tracks()
display(spots_df.head())
Downloading file 'sample_data.csv' from 'https://raw.githubusercontent.com/yfukai/laptrack/97943e8b61a5fa6e9cdabf2968037c4b1f8cbf32/docs/examples/sample_data.csv' to '/home/docs/.cache/laptrack'.
| frame | position_x | position_y | |
|---|---|---|---|
| 0 | 0 | 178.412575 | 185.188661 |
| 1 | 1 | 236.963642 | 219.971473 |
| 2 | 1 | 185.175899 | 185.188661 |
| 3 | 2 | 190.006845 | 182.483331 |
| 4 | 2 | 190.200083 | 188.280466 |
Tracking
Initializing LapTrack object
First, initialize the LapTrack object with parameters.
[4]:
max_distance = 15
lt = LapTrack(
track_dist_metric="sqeuclidean", # The similarity metric for particles. See `scipy.spatial.distance.cdist` for allowed values.
splitting_dist_metric="sqeuclidean",
merging_dist_metric="sqeuclidean",
# the square of the cutoff distance for the "sqeuclidean" metric
track_cost_cutoff=max_distance**2,
splitting_cost_cutoff=max_distance**2, # or False for non-splitting case
merging_cost_cutoff=max_distance**2, # or False for non-merging case
)
Example 1: using predict_dataframe to track pandas DataFrame coordinates
predict_dataframe is the easiest option when you have the coordinate data in pandas DataFrame.
[5]:
track_df, split_df, merge_df = lt.predict_dataframe(
spots_df,
coordinate_cols=[
"position_x",
"position_y",
], # the column names for the coordinates
frame_col="frame", # the column name for the frame (default "frame")
only_coordinate_cols=False, # if False, returned track_df includes columns not in coordinate_cols.
# False will be the default in the major release.
)
track_df is the original dataframe with additional columns “track_id” and “tree_id”.
The track_id is a unique id for each track segments without branches. A new id is assigned when a splitting and merging occured.
The tree_id is a unique id for each “clonal” tracks sharing the same ancestor.
[6]:
keys = ["position_x", "position_y", "track_id", "tree_id"]
display(track_df[keys].head())
| position_x | position_y | track_id | tree_id | ||
|---|---|---|---|---|---|
| frame | index | ||||
| 0 | 0 | 178.412575 | 185.188661 | 0 | 0 |
| 1 | 0 | 236.963642 | 219.971473 | 1 | 1 |
| 1 | 185.175899 | 185.188661 | 0 | 0 | |
| 2 | 0 | 190.006845 | 182.483331 | 2 | 0 |
| 1 | 190.200083 | 188.280466 | 3 | 0 |
split_df is a dataframe for splitting events with the following columns: - “parent_track_id” : the track id of the parent - “child_track_id” : the track id of the parent
[7]:
display(split_df)
| parent_track_id | child_track_id | |
|---|---|---|
| 0 | 0 | 2 |
| 1 | 0 | 3 |
merge_df is a dataframe for merging events with the following columns: - “parent_track_id” : the track id of the parent - “child_track_id” : the track id of the parent
[8]:
display(merge_df)
| parent_track_id | child_track_id | |
|---|---|---|
| 0 | 3 | 5 |
| 1 | 4 | 5 |
We can display tracks in napari as follows:
[9]:
# Does not work in Colab
#
# import napari
# v = napari.Viewer()
# v.add_points(spots_df[["frame", "position_x", "position_y"]])
# track_df2 = track_df.reset_index()
# v.add_tracks(track_df2[["track_id", "frame", "position_x", "position_y"]])
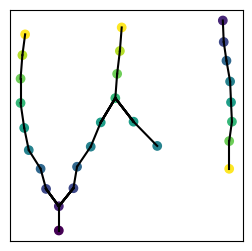
Plotting tracks in matplotlib
[10]:
plt.figure(figsize=(3, 3))
frames = track_df.index.get_level_values("frame")
frame_range = [frames.min(), frames.max()]
k1, k2 = "position_y", "position_x"
keys = [k1, k2]
def get_track_end(track_id, first=True):
df = track_df[track_df["track_id"] == track_id].sort_index(level="frame")
return df.iloc[0 if first else -1][keys]
for track_id, grp in track_df.groupby("track_id"):
df = grp.reset_index().sort_values("frame")
plt.scatter(df[k1], df[k2], c=df["frame"], vmin=frame_range[0], vmax=frame_range[1])
for i in range(len(df) - 1):
pos1 = df.iloc[i][keys]
pos2 = df.iloc[i + 1][keys]
plt.plot([pos1[0], pos2[0]], [pos1[1], pos2[1]], "-k")
for _, row in list(split_df.iterrows()) + list(merge_df.iterrows()):
pos1 = get_track_end(row["parent_track_id"], first=False)
pos2 = get_track_end(row["child_track_id"], first=True)
plt.plot([pos1[0], pos2[0]], [pos1[1], pos2[1]], "-k")
plt.xticks([])
plt.yticks([])
[10]:
([], [])
Example 2: using predict to track frame-wise-organized coordinates to make networkx tree
predict_dataframe is a thin wrapper of the predict function, the core tracking function of the LapTrack object.
One can directly use this function with the input of the frame-wise coordinate list and the output of networkx DiGraph object representing the lineage tree.
[11]:
frame_max = spots_df["frame"].max()
coords = []
for i in range(frame_max):
df = spots_df[spots_df["frame"] == i]
coords.append(df[["position_x", "position_y"]].values)
The input variable coords should be organized as the frame-wise list of the point coordinates.
The coordinate dimension is (particle, dimension).
[12]:
display(coords[:5])
[array([[178.41257464, 185.18866074]]),
array([[236.9636424 , 219.97147327],
[185.1758993 , 185.18866074]]),
array([[190.00684549, 182.48333087],
[190.20008333, 188.2804663 ],
[230.97326914, 220.16471112]]),
array([[195.61074306, 181.32390379],
[196.1904566 , 189.05341768],
[225.75584726, 220.74442466]]),
array([[200.82816494, 178.81181177],
[201.79435418, 191.9519854 ],
[201.98759203, 206.05834826],
[219.95871183, 221.51737605]])]
predict function generates a networkx DiGraph object from the coordinates
[13]:
track_tree = lt.predict(
coords,
)
The returned track_tree is the connection between spots, represented as ((frame1,index1), (frame2,index2)) …
For example, ((0, 0), (1, 1)) means the connection between [178.41257464, 185.18866074] and [185.1758993 , 185.18866074] in this example.
[14]:
for edge in list(track_tree.edges())[:5]:
print(edge)
((0, 0), (1, 1))
((1, 0), (2, 2))
((1, 1), (2, 0))
((1, 1), (2, 1))
((2, 0), (3, 0))